
The influence of genetic risk and lifestyle on the development of coronary artery disease
In the past years, almost 100 genomic loci have been identified to be genome-wide significantly associated with coronary artery disease (CAD) (1). The alleles conferring the risk at these loci are often frequently found in the European population, such that each person carries one or two of these risk alleles per locus. Thus, every individual carries a given number of risk alleles, some more—resulting in a high genetic risk score for CAD—and some less. Khera and colleagues asked the question whether a healthy lifestyle can modulate the genetic risk. They analyzed the additive effects of genetic and lifestyle factors in three prospective cohorts and one cross-sectional study to assess the event rate of coronary artery disease in over 55.000 participants. The assessment was based on genetic data from the Atherosclerosis Risk in Communities (ARIC) study (2), the Women’s Genome Health Study (WGHS) (3), the Malmö Diet and Cancer Study (MDCS) (4), and the BioImage Study (5). The median follow-up time comprised almost 20 years.
The genetic risk was evaluated using a polygenic scoring system based on 50 single nucleotide polymorphisms (SNPs) known to be associated with coronary artery disease multiplied by the predicted effect size of each risk allele (6-14). Based on the number of risk alleles per individual and their respective effect sizes the populations were divided into quintiles of the genetic score. Individuals in the lowest quintile in the distribution curve were defined having a low genetic risk, the highest quintile as having a high genetic risk, and the 60% in between were defined as being at intermediate risk. The influence of lifestyle was graded by the prevalence of four common factors—the absence of obesity, no current smoking, regular physical activity and a healthy diet. Participants reaching three or four healthy lifestyle factors were defined as having a favorable lifestyle risk, two healthy lifestyle factors were defined as at intermediate risk, and reaching one or less healthy lifestyle factors as unfavorable lifestyle risk. The study quantified the extent to which an increased genetic risk can be offset by a healthy lifestyle (15).
Primary end point of the study was the composite of myocardial infarction, coronary revascularization, and death from coronary cause. In the BioImage study, standardized coronary-artery calcification scores at baseline were also available (16). In the statistical analysis, hazard ratios and Cox-regression models were used to calculate a 10-year event rate, according to genetic and lifestyle risk.
The authors described a continuous gradient for the genetic risk regarding coronary events in all three prospective cohorts over almost 20 years of follow-up, i.e., individuals at high genetic risk score displayed a significant, 1.91-fold increased relative risk to experience coronary events compared to those at low genetic risk. Individuals with an intermediate genetic risk score also exhibited intermediate risk for coronary events in all three prospective cohorts. Genetic risk categories were independent of cardiometabolic risk factors, except for higher LDL cholesterol and family history of coronary events, which were related to a higher genetic risk score.
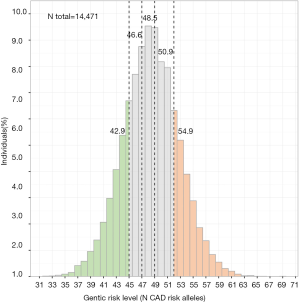
In order to illustrate the absolute differences in the genetic risk score we analyzed 14,471 control individuals of European ancestry (Figure 1). The data in this population reveal a Gaussian distribution with an average of about 43 risk alleles in the quintile of low genetic risk and about 55 risk alleles in the quintile of high genetic risk (6,9-11,13,17,18). Thus, the quintile with the low genetic risk score is not free of risk alleles! Rather, no individual—carrying 0–2 risk alleles at each locus—in the population was found to have less than 30 risk alleles at the 56 loci. In other words, even subjects in the “low genetic risk group” carry a substantial genetic risk despite having a substantially lower number of risk alleles and thus less coronary events than those in the “high genetic risk group” as studied by Khera et al.
All healthy lifestyle factors were themselves associated with a reduced incidence of coronary events. On the other hand, participants with unfavorable lifestyle were at significantly increased risk for coronary events compared to those with favorable lifestyle.
The authors next studied subgroups analyzing by which extent a healthy lifestyle can neutralize a high genetic risk and vice versa. Remarkably, the relative risk reduction of coronary artery disease was almost 50% in participants at high genetic risk but a favorable lifestyle compared to those who had both, a high genetic risk and an unfavorable lifestyle.
On the other hand it should be mentioned that individuals at high genetic risk but with a favorable lifestyle display a higher standardized 10-year coronary event rate than those individuals at low genetic risk but with an unhealthy lifestyle. In other words: a lazy, smoking individual gifted with a low number of CAD risk alleles is at lower risk of coronary events than a person that does not smoke and is physically active but carrying the burden of a high number of CAD risk alleles (Figure 2).
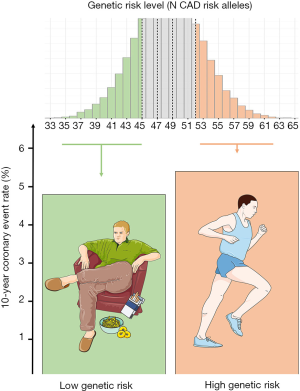
The results represent a reliable scale for a white European-American collective. For black participants, at least an association between polygenic risk score and coronary disease risk was confirmed. Whether these findings will also be applicable to other ethnic backgrounds remains to be proven. The authors described several limitations combining the data of all three prospective cohorts. Main limitation might be a different acquisition of lifestyle factors at baseline and the compliance to a healthy lifestyle that ruled out a coherent randomization. Nevertheless, the results prove a strong association between lifestyle and coronary events. In addition, lifestyle and genetic factors independently influence the risk of coronary events.
The results of Khera and colleagues underline the importance and significance of genome wide association studies (GWAS). Over the last decade large human GWAS have robustly mapped thousands of loci for various common diseases. The identification of multiple CAD loci in large GWAS presents an opportunity to quantify individual genetic risk using polygenic scores of DNA sequence polymorphisms. In general, the identified loci individually exhibit a very modest effect size and together explain solely a fraction of the heritability of CAD. In case of CAD, published meta-analyses have now identified a total of 57 loci that meet a Bonferroni-adjusted threshold level of genome-wide significance, almost the same number is about to be published and about hundreds of additional loci exhibit suggestive association (13,14,17,19-21).
Further stratification of the genetic risk in the quintile of low genetic risk would be of peculiar interest. Regarding almost 50 risk alleles, known to a have an effect on the development of coronary artery disease, a hypothetical question arises: what would be the lowest genetic risk possible (0 out of 50 risk alleles)? Although this would apply only for very few individuals, it would make up the full picture.
The unique characteristic of this study is the large number of participants and at the same time the length of follow-up. In combination, this study offers an enormous significance though there are several limitations. The results emphasize an already existing consensus of cardiologists world-wide: subjects with a distinct lifestyle behavior are at increased risk of cardiovascular events. Given the enormous genetic effects as shown by Khera et al and others before it will be interesting to resolve the question whether genetic testing should become part of the global risk assessment as well (22). Moreover the paper of Khera clarifies that the incidence of coronary artery disease can be influenced by a healthy lifestyle independent from the genetic predisposition. To reduce coronary events to a minimum, individuals at all levels of genetic risk should be motivated to permanently reduce their lifestyle risk factors.
Acknowledgments
Individual level data of Figure 1 were obtained from the German Myocardial Infarction Family Studies (GerMIFS) I, II, III (KORA), IV, V, VI, Myocardial Infarction Genetics Consortium (MIGen), the Cardiogenics Consortium, and the Wellcome Trust Case Control Consortium (WTCCC). All subjects in all studies were Caucasians of European origin and gave written informed consent before participating. We thank all participants. Figure 2 contains modified image material available at Servier Medical Art under a Creative Commons Attribution 3.0 Unported License.
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, Journal of Public Health and Emergency. The article did not undergo external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jphe.2017.03.06). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Kessler T, Vilne B, Schunkert H. The impact of genome-wide association studies on the pathophysiology and therapy of cardiovascular disease. EMBO Mol Med 2016;8:688-701. [Crossref] [PubMed]
- The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. The ARIC investigators. Am J Epidemiol 1989;129:687-702. [Crossref] [PubMed]
- Ridker PM, Chasman DI, Zee RY, et al. Rationale, design, and methodology of the Women's Genome Health Study: a genome-wide association study of more than 25,000 initially healthy american women. Clin Chem 2008;54:249-55. [Crossref] [PubMed]
- Berglund G, Elmstahl S, Janzon L, et al. The Malmo Diet and Cancer Study. Design and feasibility. J Intern Med 1993;233:45-51. [Crossref] [PubMed]
- Muntendam P, McCall C, Sanz J, et al. The BioImage Study: novel approaches to risk assessment in the primary prevention of atherosclerotic cardiovascular disease--study design and objectives. Am Heart J 2010;160:49-57.e1. [Crossref] [PubMed]
- Samani NJ, Erdmann J, Hall AS, et al. Genomewide association analysis of coronary artery disease. N Engl J Med 2007;357:443-53. [Crossref] [PubMed]
- Helgadottir A, Thorleifsson G, Manolescu A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science 2007;316:1491-3. [Crossref] [PubMed]
- McPherson R, Pertsemlidis A, Kavaslar N, et al. A common allele on chromosome 9 associated with coronary heart disease. Science 2007;316:1488-91. [Crossref] [PubMed]
- Kathiresan S, Voight BF, Purcell S, et al. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet 2009;41:334-41. [Crossref] [PubMed]
- Erdmann J, Grosshennig A, Braund PS, et al. New susceptibility locus for coronary artery disease on chromosome 3q22.3. Nat Genet 2009;41:280-2. [Crossref] [PubMed]
- Peden JF, Hopewell JC, Saleheen D, et al. A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat Genet 2011;43:339-44. [Crossref] [PubMed]
- Butterworth AS, Braund PS, Farrall M, et al. Large-scale gene-centric analysis identifies novel variants for coronary artery disease. PLoS Genet 2011;7:e1002260 [Crossref] [PubMed]
- Deloukas P, Kanoni S, Willenborg C, et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet 2013;45:25-33. [Crossref] [PubMed]
- Nikpay M, Goel A, Won HH, et al. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet 2015;47:1121-30. [Crossref] [PubMed]
- Khera AV, Emdin CA, Drake I, et al. Genetic Risk, Adherence to a Healthy Lifestyle, and Coronary Disease. N Engl J Med 2016;375:2349-58. [Crossref] [PubMed]
- Baber U, Mehran R, Sartori S, et al. Prevalence, impact, and predictive value of detecting subclinical coronary and carotid atherosclerosis in asymptomatic adults: the BioImage study. J Am Coll Cardiol 2015;65:1065-74. [Crossref] [PubMed]
- Schunkert H, Konig IR, Kathiresan S, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet 2011;43:333-8. [Crossref] [PubMed]
- Burton PR, Clayton DG, Cardon LR, et al. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 2007;447:661-78. [Crossref] [PubMed]
- McPherson R, Tybjaerg-Hansen A. Genetics of Coronary Artery Disease. Circ Res 2016;118:564-78. [Crossref] [PubMed]
- Peden JF, Hopewell JC, Saleheen D, et al. A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat Genet 2011;43:339-44. [Crossref] [PubMed]
- Abraham G, Havulinna AS, Bhalala OG, et al. Genomic prediction of coronary heart disease. Eur Heart J 2016;37:3267-78. [Crossref] [PubMed]
- Hughes MF, Saarela O, Stritzke J, et al. Genetic markers enhance coronary risk prediction in men: the MORGAM prospective cohorts. PLoS One 2012;7:e40922 [Crossref] [PubMed]
Cite this article as: von Scheidt M, Kessler T, Zeng L, Schunkert H. The influence of genetic risk and lifestyle on the development of coronary artery disease. J Public Health Emerg 2017;1:40.


