
IRGB10 supplies bacterial ligands to activate AIM2 and NLRP3 inflammasomes
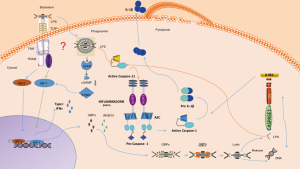
Recently, Man et al. [2016] published an elegant study in which they showed that immunity-related GTPase family member b10 (IRGB10; an interferon-inducible protein) localizes on the cell membrane of bacteria, damages the membrane, and then releases the hidden ligands from the phagosome to the host-cytoplasm to be sensed by both the absent in melanoma 2 (AIM2) and the noncanonical NLR family pyrin domain-containing (NLRP) inflammasomes (1).
At the precise moment when a pathogen interacts with the host, many mechanisms begin to function in order to eliminate the pathogen. Thus, innate immunity and the adaptive immune system play essential roles in the elimination or control of the pathogen.
Intracellular bacteria are masters of hijacking the cellular machinery; however, the infected cells respond rapidly and at an early step in the infection process. Thus, when bacteria enter the phagosome, many sensors will interact with the pathogen, then if the bacteria are able to escape into the cytoplasm, within the cytoplasm, the bacterial pathogen-associated molecular patterns (PAMPs) or damage-associated molecular patterns (DAMPs) are in contact with a host of cytosolic pattern recognition receptors (PRRs), such as NOD-like receptors (NLRs) and AIM-2 like receptors (ALRs). Then, the PRRs stimulate a pro-inflammatory and antimicrobial transcriptional response through both; phosphorylated nuclear factor kappa B (NFκB) or interferon-regulatory factors (IRFs) once they are translocated into nucleus. Assemble of a multiprotein complex called the inflammasome, which is composed of a sensor molecule (e.g., NLRP3 or AIM2), and apoptosis speck-like protein (ASC) results in activation of the inflammasome platform in response to microbial or danger signals, such as nucleic acids or lipopolysaccharide (LPS). This then leads to cleavage of pro-caspase-1 and the subsequent maturation of interleukin (IL)-1B and IL-18 (2). However, some microbes, such as Francisella novicida, evade this recognition and escape the phagosomal vacuole. These microbes have also evolved strategies to synthesize a modified LPS that cannot be sensed by caspase-11.
Recently, Man et al. established that the immunity-related GTPase family member b10 (IRGB10; an interferon-inducible protein) is involved in the damage of the cell membrane of bacteria, that facilitates the liberation of hidden ligands contained in the phagophore to target innate immune receptors (1). The first response against intracellular pathogens is mediated by IFNs molecules that are one of the early mechanisms of host that contributes determinately to eliminate pathogens. The members of IFNs family are critical, since they are engaged in the induction of the expression of more than 2,000 genes involved in the control of infection, among them, the big family of interferon-inducible GTPases that include both, the immunity-related GTPases (IRGBs) and guanylate-binding proteins (GBPs) (3).
GBP is a family of proteins involved in the recruitment of different molecules with antimicrobial activity targeting phagosomes that contain intracellular pathogens. Thus, the recognition of intracellular LPS requires bacterial escape from the vacuole. This process is also promoted by IFN-inducible GTPase-mediated lysis of bacteria contained within a vacuole, resulting in LPS-dependent, non-canonical activation of inflammatory caspases. As example, GBP-5 has been shown to have the ability to trigger the assembly of NLRP3 and AIM2 inflammasomes through flagellin and cytosolic DNA (4).
The mechanism of this phenomenon was addressed in two excellent papers published simultaneously by Man et al. and Meunier et al. in 2015. Their studies demonstrated the recruitment of GBP2 and GBP5 to bacterial membranes, followed by bacterial lysis; this process caused DNA to leak into the cytoplasm to be detected by the inflammasome sensor AIM2 (5,6). Besides, the secretion of type I INFs involves the DNA sensor cyclic GMP-AMP synthase (cGAS) via the adaptor stimulator of IFN genes (STING), which further induces type I IFN-dependent expression of IFN regulatory factor 1 (IRF1) and triggers the expression of GBPs. They then used siRNA screening for nearly 500 IFN-simulated genes to screen more than 400 ISGs and found that GBP2 and GBP5 were key AIM2 activators; indeed, silencing the expression of the genes encoding GBP2 and GBP5 compromised inflammasome responses to bacterial infection (5,6).
Thus, the IFN-inducible genes (ISGs) regulated by IRF1 participate in the activation of AIM2 for the control of F. tularensis. In this new report, Man et al. identified another important molecule regulated by IRF1, i.e., IRGB10, which supplies ligands from bacteria that may participate in NLRP3 activation. Additionally, they validated the dependency of GBPchr3 proteins on the colocalization of intracellular bacteria, such as F. tularensis (1).
The roles of different IRGs have been demonstrated previously in a wide variety of intracellular bacteria, such as Mycobacterium, Salmonella, and Listeria, in which IRGs have been shown to enhance phagosome maturation and induction of autophagy or encourage the destruction of vacuoles that contain the pathogens (3). Interestingly, a finding showed the ectopic expression of IRGB10 in the absence of IFNs was sufficient to block the intracellular growth of Chlamydia trachomatis, in contrast to C. muridarum, which evades the IRGB10 host response (7).
Man et al. elucidates the molecular mechanisms through which the IRGB10 protein mediates resistance in a model with F. tularensis. They showed that cells lacking IRGB10 infected with F. novicida failed to trigger AIM2 activation. However, IRGB10 was found dispensable for activation of AIM2 by transfected poly (dA:dT) and plasmid DNA. Similar to caspase-1 cleavage, IL-1b secretion was possible after direct LPS treatment. Thus, IRGB10 contributed to the activation of the noncanonical inflammasome upon infection with LPS-containing Escherichia coli (E. coli) and C. rodentium. The authors further demonstrated through structural analysis that IRGB10 penetrated the E. coli membrane and could use vesicles as a cellular transport mechanism to be delivered through the bacterial membrane. Moreover, bioinformatics analysis further identified myristoylation motifs and transmembrane amphipathic alpha helixes in IRGB10 that may be involved in membrane attachment, permeabilization, and disruption, ultimately leading to DNA leakage into the cytoplasm. Amphipathic helices are characteristics of microbial peptides that allow them to induce membrane permeabilization, loss of membrane potential, and membrane disruption (8). Notable, IRGB10 has also been shown to oligomerize, insert, and disrupt the membrane in C. trachomatis through its myristoylation motif and amphipathic helix (9). Furthermore, Man et al. elucidated the interactions among bacterial DNA, cellular IRGB10/GBP5, and bacterial LPS through super-resolution microscopy. Their results suggested that LPS surrounds the layer of IRGB10 and GBP5, which further surrounds bacterial DNA.
In vivo, IRGB10 was found to act against F. novicida and protect the host as the susceptibility of IRGB10-/- or GBPchr3-/- mice to F. novicida infections was significantly increased compared with that of wild-type animals. Thus, these findings have provided insights into the molecular mechanism through which innate immune receptors assist in the defense against microbes (1).
As with all new findings, some pressing questions are raised and need to be addressed in future investigations. A previous report showed the dependency of cGAS/STING in the ability of IRGB10/GBPs to access to cytosolic DNA (10). Later, the requirement of the cGAS-dependent sequential IFN-IRF1-GBP cascade for bacterial lysis to expose cytosolic bacterial DNA was demonstrated (5). The current study by Man et al. suggests that IRGB10/GBPs is required for the same process. Like AIM2, cGAS is also a DNA sensor. Therefore, it is unclear how cGAS gains access to bacterial DNA before IRGB10/GBP-dependent bacterial lysis occurs. One possibility could be the high sensitivity of cGAS to the F. tularensis DNA within the cytosol, whereas AIM2 requires the induction and activation of GBPs to liberate greater quantities of ligand. Another possibility could be the contribution of other DNA sources. In this respect, several reports showed that mitochondrial DNA that is liberated into cytoplasm upon cellular infection with intracellular pathogens, could be an alternative mechanism of cGAS to enhance the innate immune responses (11).
Man et al. identified molecular mechanisms that could provide a paracrine signal when GBPs and IRGB10 are expressed in surrounding cells through cGAS-activated cells (see Figure 1). This work confirmed once again that the IRG system is a powerful resistance mechanism against some intracellular pathogens in mammals.
Many interesting papers regarding to the members of the family of IFN-inducible GTPases and its role in the innate immunity against intracellular pathogens and their concerted function have been published recently. However many studies are required to elucidate the mechanisms of action of IRG to shed light on the orchestrated function of the other molecules cGAS-STING/GBP/IRGB10-raised along with these study to understand the dependent cytosolic immune mechanisms.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, Journal of Public Health and Emergency. The article did not undergo external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jphe.2017.02.07). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Man SM, Karki R, Sasai M, et al. IRGB10 Liberates Bacterial Ligands for Sensing by the AIM2 and Caspase-11-NLRP3 Inflammasomes. Cell 2016;167:382-396.e17. [Crossref] [PubMed]
- Broz P, Dixit VM. Inflammasomes: mechanism of assembly, regulation and signalling. Nat Rev Immunol 2016;16:407-20. [Crossref] [PubMed]
- Meunier E, Broz P. Interferon-inducible GTPases in cell autonomous and innate immunity. Cell Microbiol 2016;18:168-80. [Crossref] [PubMed]
- Kim BH, Shenoy AR, Kumar P, et al. Supporting Online Material for A Family of IFN-γ–Inducible 65-kD GTPases Protects Against Bacterial Infection. Science 2011;332:
- Man SM, Karki R, Malireddi RK, et al. The transcription factor IRF1 and guanylate-binding proteins target activation of the AIM2 inflammasome by Francisella infection. Nat Immunol 2015;16:467-75. [Crossref] [PubMed]
- Meunier E, Wallet P, Dreier RF, et al. Guanylate-binding proteins promote activation of the AIM2 inflammasome during infection with Francisella novicida. Nat Immunol 2015;16:476-84. [Crossref] [PubMed]
- Coers J, Bernstein-Hanley I, Grotsky D, et al. Chlamydia muridarum evades growth restriction by the IFN-gamma-inducible host resistance factor Irgb10. J Immunol 2008;180:6237-45. [Crossref] [PubMed]
- Dathe M, Wieprecht T. Structural features of helical antimicrobial peptides: their potential to modulate activity on model membranes and biological cells. Biochim Biophys Acta 1999;1462:71-87. [Crossref] [PubMed]
- Haldar AK, Saka HA, Piro AS, et al. IRG and GBP host resistance factors target aberrant, "non-self" vacuoles characterized by the missing of "self" IRGM proteins. PLoS Pathog 2013;9:e1003414 [Crossref] [PubMed]
- Xiao TS, Fitzgerald KA. The cGAS-STING pathway for DNA sensing. Mol Cell 2013;51:135-9. [Crossref] [PubMed]
- Wiens KE, Ernst JD. The Mechanism for Type I Interferon Induction by Mycobacterium tuberculosis is Bacterial Strain-Dependent. PLoS Pathog 2016;12:e1005809 [Crossref] [PubMed]
Cite this article as: Shrivastava G, Gutierrez-Castañeda B, García-Cordero J, León-Juárez M, Cedillo-Barrón L. IRGB10 supplies bacterial ligands to activate AIM2 and NLRP3 inflammasomes. J Public Health Emerg 2017;1:31.


